
Low expression of KLRB1 predicts poor survival outcomes and is associated with immune infiltration in breast cancer
Highlight box
Key findings
• KLRB1 is a potential predictive tumor marker for breast cancer (BRCA) patients.
What is known and what is new?
• BRCA seriously endangers women’s health. It is the most commonly diagnosed cancer in women, and ranks second among the causes of cancer-related deaths in women.
• The high expression of KLRB1 is associated with prognostic significance.
What is the implication, and what should change now?
• By analyzing the expression and function of KLRB1 gene, we expect to find its important role in the occurrence and development of BRCA, and provide new strategies and methods for the treatment of BRCA.
• Report about implications and actions is needed.
Introduction
Globally, the burden of breast cancer (BRCA) is increasing. This disease stands as the most prevalent type of cancer in females and a leading contributor to cancer-related mortality (1,2). While advancements in early diagnosis and comprehensive treatment approaches have led to improved prognoses for individuals with BRCA, the 5-year overall survival (OS) rate remains below 20% when metastasis is present (3,4). Hence, there is an urgent need to identify biological markers associated with BRCA prognosis.
Recent research has indicated a potential close association between the KLRB1 gene and the onset and progression of BRCA (5). KLRB1 gene is a cell surface molecule belonging to the C-type lectin family and has a variety of biological functions (6). In the immune response, the KLRB1 gene can inhibit the activation of T lymphocytes, thus playing an immunomodulatory role (7). Multiple investigations have explored the expression and role of the KLRB1 gene in BRCA. These studies have frequently revealed a reduced expression or deletion of the KLRB1 gene within BRCA cells, showcasing a strong association with the invasive and metastatic tendencies of tumors (8,9). Additionally, the KLRB1 gene can affect the biological behavior of BRCA cells by regulating cell apoptosis, cell cycle, cell invasion, and other processes (10). In addition, recent research also found that the KLRB1 gene may be associated with chemotherapy resistance of BRCA, offering a novel potential target for BRCA treatment (11). However, the role of the KLRB1 gene in BRCA remains uncertain. Therefore, this study aims to explore the mechanism and potential therapeutic targets of the KLRB1 gene in BRCA.
As high-throughput sequencing technology has advanced, the generation of extensive omics data has become feasible (12,13,14). The The Cancer Genome Atlas (TCGA)-BRCA gene can help elucidate the causes and prognosis of cancer. This research analyzed the transcriptional levels and prognosis-predictive value of KLRB1 using the data acquired from TCGA-BRCA. Furthermore, the biological mechanism of KLRB1 was investigated using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, and the relationship between KLRB1 and immune infiltration levels was assessed. Additionally, quantitative polymerase chain reaction (qPCR), immunohistochemistry (IHC), and Cell Counting Kit-8 (CCK8) experiments provided validation for our findings. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1231/rc).
Methods
Data processing
Expression profiles of 1,109 BRCA tissues and 113 adjoining tissues were retrieved from TCGA. Subsequently, clinicopathological features and predictive data of the individuals were subjected to further screening. RNA-seq (RNA sequencing) data in transcripts per kilobase million (TPM) format from TCGA were uniformly processed. The expression of KLRB1 was evaluated using TCGA. To assess the level of KLRB1 expression in the pan-cancer, extracted data from UCSC Xena were assessed (https://xenabrowser.net/datapages/).
Patients and tissues
Ten pairs of BRCA samples and their matched non-tumor tissues were acquired from Liaoning Cancer Hospital. Every participant provided written informed consent. The approval for this research was granted by the ethics committee of Liaoning Cancer Hospital (20210621) and the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The BRCA tissues were collected post-surgery, immediately frozen in liquid nitrogen, and then preserved at −80 ℃ for qPCR analysis.
Gene enrichment analysis
By utilizing the transcriptional sequence from TCGA, the study employed GO and KEGG analyses to determine the genes and pathways associated with KLRB1. The expression data were classified into groups with high and low KLRB1 expression (R “clusterProfiler”).
Immune cell infiltration
To evaluate the relative abundance of infiltrating immune cells in tumor tissues, single sample gene set enrichment analysis (ssGSEA) was conducted. The infiltration levels of immune cells in BRCA expression data were assessed utilizing R “gsva” and an immune data set, including 24 immune cells.
Survival and prognosis analysis
The “survival” graph of KLRB1 was utilized to derive the OS. A division threshold of 50% was chosen as a critical value to divide the cohort into high- and low-expression groups. To examine the prognostic significance of KLRB1 in individuals with BRCA, the “roc” function from the R package was employed for analysis, and visualization was performed using the “ggplot2” package.
Cell culture and transfection
The MCF10A cell line, MCF7 cell line and MDAMB231 cell line was acquired from the Chinese Academy of Sciences and cultured in minimum essential medium (MEM) comprising 10% fetal bovine serum (FBS; GIBCO, Waltham, MA, USA) and 1% penicillin-streptomycin. Cells were cultured in a humidified incubator under 5% CO2 at 37 ℃. One day prior to transfection, MCF7 and MDAMB231 cells were cultured in six-well plates to achieve 50–60% confluence. Transfection of the KLRB1-targeted pEZ-M03 vector was carried out using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) as per the provided guidelines.
RNA isolation and qPCR analysis
The extraction of tissue RNA was carried out using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Subsequently, the extracted RNA was converted into complementary DNA (cDNA) using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA, USA). The qPCR technique quantifies DNA during each cycle of the PCR reaction through real-time fluorescence measurements. The analyses were conducted using SYBR-Green (Takara, Otsu, Shiga, Japan) as the detection dye, with GAPDH serving as the internal control. The primers employed were as follows: KLRB1 forward primer, 5'-AATTTGCCCTGAAACTTAGCTG-3'; reverse, 5'-GGATGTCACTGAAACACTCAAC-3'. GAPDH forward primer, 5'-GTCTCCTCTGACTTCAACAGCG-3'; reverse, 5'-ACCACCCTGTTGCTGTAGCCAA-3'. The qPCR value was calculated using the 2-delta delta Ct method. We set the Ct value to 15-35, and Ct values not in this range will be excluded.
IHC
IHC was performed using a two-step method according to the manufacturer’s instructions (PV-9000; ZSGB-BIO, Beijing, China). BRCA samples were fixed in 10% formalin, paraffin-embedded, and sectioned into 5-µm slices. The samples were de-waxed with ethanol and blocked to inactivate the endogenous peroxidase activity. Subsequently, antigen retrieval was achieved by heating the samples in a microwave, followed by cooling to room temperature. Blocking was performed using goat serum for 30 minutes at 37 ℃. The samples were then incubated overnight at 4 ℃ with rabbit anti-KLRB1 (Thermo Fisher Scientific 17-5941-82) (1:200). Afterward, incubation with horseradish peroxidase-coupled goat anti-rabbit secondary antibody (PV-9000; ZSGB-BIO, Beijing, China) was conducted at 37 ℃ for 30 minutes. The samples were then stained with 3,3'-diaminobenzidine (DAB). Cell nuclei were stained blue with hematoxylin. The sections were then dehydrated, cleared with xylene, and mounted. KLRB1 expressions were determined by IHC using the streptavidin peroxidase method, with adjacent tissues serving as the controls. The experimental procedure was performed as per the manufacturer’s instructions. Image-Pro Plus 6.0 Software (Media Cybernetics, USA) was used to analyze protein expression and perform statistical analysis of the results obtained by IHC.
Cell colony formation assay
The cells were planted at a density of 1×103/mL in each well of the 6-well plates, with 2 mL of MEM medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. Single-cell-derived clones were allowed to grow for ten days. Prior to fixation, the culture was pre-cooled three times with phosphate buffered saline (PBS). The cells were then fixed with methanol for 15 minutes, stained with crystal violet for 20 minutes, and rinsed with water. The dishes were air dried, and the number of visible clones was visually counted. The colony formation rate was calculated. This entire procedure was repeated three times to ensure reproducibility.
Transwell assay
The cells were collected, resuspended in serum-free media, and introduced into the upper compartment of a Transwell membrane filter that had been coated with Matrigel (Corning) for invasion assays. To the lower compartment, we added a culture medium containing 10% FBS and either 0, 5, or 10 nM Tanespimycin as a chemoattractant. After a 36-hour incubation period, the cells were fixed with methanol, stained with 0.1% crystal violet, and then imaged and counted using an Olympus microscope (Tokyo, Japan). For the migration assay, the process was repeated for 24 hours.
Statistical analysis
The Wilcoxon rank-sum test was employed to conduct statistical analysis on the expression of KLRB1 in both the healthy and BRCA groups. Individuals were classified into two groups as per their “median” expression of KLRB1. The clinical and pathological characteristics of KLRB1 were assessed utilizing the Wilcoxon-rank sum test or Kruskal-Wallis test and logistic regression. Prognosis-predictive analysis was conducted utilizing Kaplan-Meier analysis as well as univariate and multivariate Cox analyses. The diagnostic value of differentially expressed genes (DEGs) was analyzed by generating a receiver operating characteristic (ROC) curve utilizing the “proc” package.
Results
Expression analysis of KLRB1 in pan-cancer and BRCA
Data downloaded from TCGA and Genotype-Tissue Expression (GTEX) were used to evaluate the expression of KLRB1 in 33 cancers. The results demonstrated that KLRB1 exhibited low expression in various cancer types, including BRCA, bladder urothelial carcinoma (BLCA), kidney chromophobe (KICH), colon adenocarcinoma (COAD), pancreatic adenocarcinoma (PAAD), liver hepatocellular carcinoma (LIHC), head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD), rectum adenocarcinoma (READ), lung squamous cell carcinoma (LUSC), thyroid carcinoma (THCA) and uterine corpus endometrial carcinoma (UCEC). However, the expression of KLRB1 was high in kidney renal clear cell carcinoma (KIRC), glioblastoma multiforme (GBM), and kidney renal papillary cell carcinoma (KIRP) (Figure 1A). The link between KLRB1 expression and clinical outcomes of individuals with BRCA was further investigated. Survival analysis demonstrated remarkable variations among distinct cancer types (Figure 1B-1D). Within the BRCA cohort, individuals exhibiting elevated KLRB1 levels displayed extended OS, progression-free interval (PFI), and disease-specific survival (DSS) in contrast to those with lowered KLRB1 levels. Furthermore, the evaluation of KLRB1 expression in BRCA within the TCGA database provided confirmation of its lower expression in this context (Figure 1E,1F). KLRB1 messenger RNA (mRNA) expression in human epidermal growth factor receptor 2 (HER2), Luminal A, Luminal B and triple negative breast cancer (TNBC) tissues were no significant difference (Figure 1G). And gene deletion was an important factor for KLRB1 down-regulation in BRCA (Figure 1H).
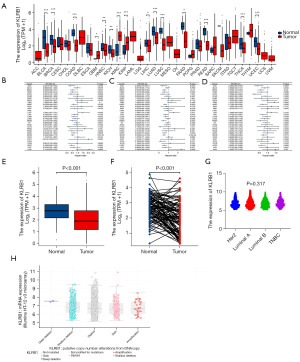
Clinical correlation of KLRB1 expression in individuals with BRCA
The clinical features and gene expression profiles of 1083 individuals with primary BRCA were acquired from the TCGA database. Individuals were classified into high (n=542) and low (n=541) KLRB1 expression groups. The aim was to examine the link between the KLRB1 expression and the clinical and pathological attributes of individuals. The analysis demonstrated a link between KLRB1 expression and M stage (P=0.043) as well as age (P<0.001), utilizing the chi-square test or Fisher’s exact test (Table 1).
Table 1
| Characteristics | Low expression of KLRB1 (n=541) | High expression of KLRB1 (n=542) | P |
|---|---|---|---|
| T stage, n (%) | 0.371 | ||
| T1 | 132 (12.2) | 145 (13.4) | |
| T2 | 321 (29.7) | 308 (28.5) | |
| T3 | 64 (5.9) | 75 (6.9) | |
| T4 | 21 (1.9) | 14 (1.3) | |
| N stage, n (%) | 0.288 | ||
| N0 | 268 (25.2) | 246 (23.1) | |
| N1 | 173 (16.3) | 185 (17.4) | |
| N2 | 52 (4.9) | 64 (6.0) | |
| N3 | 33 (3.1) | 43 (4.0) | |
| M stage, n (%) | 0.043 | ||
| M0 | 447 (48.5) | 455 (49.3) | |
| M1 | 15 (1.6) | 5 (0.5) | |
| Pathologic stage, n (%) | 0.070 | ||
| Stage I | 85 (8.0) | 96 (9.1) | |
| Stage II | 321 (30.3) | 298 (28.1) | |
| Stage III | 110 (10.4) | 132 (12.5) | |
| Stage IV | 13 (1.2) | 5 (0.5) | |
| Race, n (%) | 0.204 | ||
| Asian | 34 (3.4) | 26 (2.6) | |
| Black or African American | 97 (9.8) | 84 (8.5) | |
| White | 361 (36.3) | 392 (39.4) | |
| Age (years), n (%) | <0.001 | ||
| ≤60 | 267 (24.7) | 334 (30.8) | |
| >60 | 274 (25.3) | 208 (19.2) | |
| PR status, n (%) | 0.436 | ||
| Negative | 163 (15.8) | 179 (17.3) | |
| Indeterminate | 1 (0.1) | 3 (0.3) | |
| Positive | 346 (33.5) | 342 (33.1) | |
| ER status, n (%) | 0.165 | ||
| Negative | 107 (10.3) | 133 (12.9) | |
| Indeterminate | 1 (0.1) | 1 (0.1) | |
| Positive | 403 (38.9) | 390 (37.7) | |
| HER2 status, n (%) | 0.495 | ||
| Negative | 255 (35.1) | 303 (41.7) | |
| Indeterminate | 6 (0.8) | 6 (0.8) | |
| Positive | 80 (11.0) | 77 (10.6) | |
| Age (years), median [IQR] | 61 [51, 70] | 55 [47, 64] | <0.001 |
BRCA, breast cancer; PR, progesterone receptor; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; IQR, interquartile range.
Link between KLRB1 expression and survival prognosis of individuals with BRCA
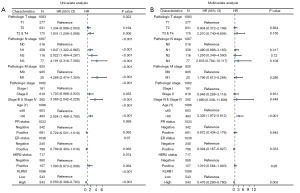
Univariate and multivariate Cox analyses were conducted to examine the impact of various factors on the OS of BRCA patients, as detailed in Table 2. In the univariate Cox analysis, KLRB1 expression (P<0.001), T3&T4 stage (P=0.006), N stage (P<0.001), M1 stage (P<0.001), pathological stage III&IV (P<0.001), and age >60 (P<0.001) were linked to the OS. The multivariate Cox model revealed an association with poor prognosis for age >60 (P<0.001), estrogen receptor (ER) status positive (P=0.033), and KLRB1 expression (P=0.003) (Table 2). Furthermore, the link between KLRB1 expression and OS, DSS, and PFI in individuals with BRCA was investigated. The Kaplan-Meier (KM) diagram illustrated that elevated KLRB1 expression was associated with a poorer prognosis for OS [hazard ratio (HR) 0.55, 95% confidence interval (CI): 0.40–0.76, P<0.001] (Figure 2A). Regarding DSS, individuals with elevated KLRB1 levels still exhibited a poorer prognosis (HR 0.51, 95% CI: 0.33–0.79, P=0.003) (Figure 2B). Similarly, for PFI, individuals with elevated KLRB1 expression also experienced a poorer prognosis (HR 0.57, 95% CI: 0.41–0.79, P<0.001) (Figure 2C). However, the low expression of KLRB1 has no significant difference in the prognosis of BRCA patients with different subtypes (Figure 2D). Furthermore, ROC curve analysis was conducted to assess the capability of differentiating BRCA tissues from healthy breast tissues as per KLRB1 expression levels. The area under the ROC curve (AUC) was 0.712 (Figure 2E). Hence, KLRB1 may become a promising prognostic biological marker for patients with BRCA.
Table 2
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | |||
| Pathologic T stage | 1,083 | 0.023 | ||||
| T1 | 277 | Reference | Reference | |||
| T2 | 631 | 1.334 (0.889–2.003) | 0.164 | 0.904 (0.372–2.199) | 0.824 | |
| T3 & T4 | 175 | 1.931 (1.208–3.088) | 0.006 | 2.210 (0.740–6.604) | 0.156 | |
| Pathologic N stage | 1,067 | <0.001 | ||||
| N0 | 516 | Reference | Reference | |||
| N1 | 358 | 1.947 (1.322–2.865) | <0.001 | 1.480 (0.686–3.193) | 0.317 | |
| N2 | 116 | 2.522 (1.484–4.287) | <0.001 | 1.265 (0.349–4.590) | 0.720 | |
| N3 | 77 | 4.191 (2.318–7.580) | <0.001 | 2.835 (0.794–10.117) | 0.108 | |
| Pathologic M stage | 925 | <0.001 | ||||
| M0 | 905 | Reference | Reference | |||
| M1 | 20 | 4.266 (2.474–7.354) | <0.001 | 1.796 (0.613–5.264) | 0.286 | |
| Pathologic stage | 1,062 | <0.001 | ||||
| Stage I | 181 | Reference | Reference | |||
| Stage II | 619 | 1.703 (0.989–2.933) | 0.055 | 0.948 (0.289–3.118) | 0.931 | |
| Stages III & IV | 262 | 3.566 (2.042–6.228) | <0.001 | 1.980 (0.338–11.606) | 0.449 | |
| Age (years) | 1,086 | <0.001 | ||||
| ≤60 | 603 | Reference | Reference | |||
| >60 | 483 | 2.024 (1.468–2.790) | <0.001 | 3.326 (1.972–5.612) | <0.001 | |
| PR status | 1,033 | 0.068 | ||||
| Negative | 342 | Reference | Reference | |||
| Positive | 691 | 0.729 (0.521–1.019) | 0.065 | 0.972 (0.434–2.176) | 0.945 | |
| ER status | 1,036 | 0.070 | ||||
| Negative | 240 | Reference | Reference | |||
| Positive | 796 | 0.709 (0.493–1.019) | 0.063 | 0.394 (0.167–0.927) | 0.033 | |
| HER2 status | 717 | 0.074 | ||||
| Negative | 560 | Reference | Reference | |||
| Positive | 157 | 1.593 (0.973–2.609) | 0.064 | 1.019 (0.564–1.840) | 0.950 | |
| KLRB1 | 1,086 | <0.001 | ||||
| Low | 543 | Reference | Reference | |||
| High | 543 | 0.550 (0.396–0.763) | <0.001 | 0.476 (0.290–0.780) | 0.003 | |
OS, overall survival; BRCA, breast cancer; CI, confidence interval; PR, progesterone receptor; ER, estrogen receptor; HER2, human epidermalgrowth factor receptor 2.
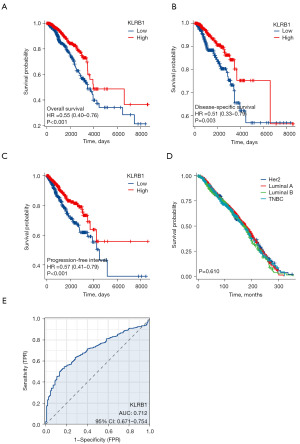
Survival analysis
Subsequently, univariate and multivariate analyses were conducted. In the former, reduced expression of T3 and T4 within the T stage, N1, N2, and N3 within the N stage, M1 within the M stage, stages 3 and 4 within the pathological stage, age >60, and KLRB1 were linked to OS. The subsequent multivariate analysis indicated independent risk factors. Specifically, age >60, ER status Positive, and reduced expression of KLRB1 were identified as independent predictive factors for OS among inpatients with BRCA (Table 2 and Figure 3).
Enrichment analysis of KLRB1-related genes
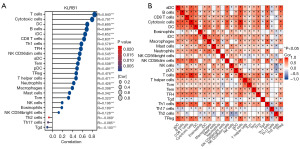
A total of 24,593 genes exhibited differential expression between the groups characterized by low and high KLRB1 expression levels, including 8 genes with lowered expression levels and 20 genes with elevated expression levels (adjusted P value <0.05, |log2 fold change (FC)|>3) (Figure 4A and table available at https://cdn.amegroups.cn/static/public/tcr-23-1231-1.xlsx). The results of GO and KEGG joint analysis of DEGs demonstrated that it was mainly enriched in signaling receptor activator activity, lymphocyte proliferation, mononuclear cell proliferation, leucocyte proliferation, receptor-ligand activity, immunoglobulin binding and hematopoietic cell lineage signaling pathways (Figure 4B and Table 3). Moreover, the GSEA of the detected DEGs revealed several immune-related biological processes. These included KEGG OLFACTORY TRANSDUCTION, REACTOME OLFACTORY SIGNALING PATHWAY, NABA SECRETED FACTORS, KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION and KEGG SYSTEMIC LUPUS ERYTHEMATOSUS (Figure 4C). Next, the correlation between the top 10 upregulated and downregulated DEGs and KLRB1 was examined, revealing significant associations between the majority of DEGs and KLRB1 (Figure 4D-4E).
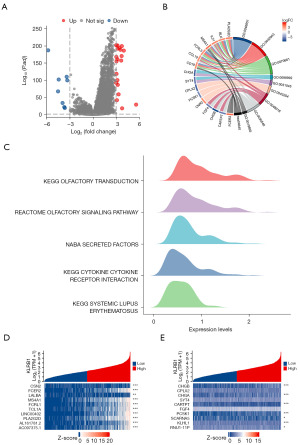
Table 3
| Ontology | ID | Description | Gene ratio | Bg ratio | P value | P.adjust | Z score |
|---|---|---|---|---|---|---|---|
| BP | GO:0046651 | Lymphocyte proliferation | 7/23 | 296/18,800 | 4.41e−08 | 1.91e−05 | 2.6457513 |
| BP | GO:0032943 | Mononuclear cell proliferation | 7/23 | 300/18,800 | 4.84e−08 | 1.91e−05 | 2.6457513 |
| BP | GO:0070661 | Leukocyte proliferation | 7/23 | 330/18,800 | 9.28e−08 | 2.36e−05 | 2.6457513 |
| CC | GO:0098992 | Neuronal dense core vesicle | 2/23 | 13/19,594 | 0.0001 | 0.0049 | −1.4142136 |
| CC | GO:0031045 | Dense core granule | 2/23 | 26/19,594 | 0.0004 | 0.0101 | −1.4142136 |
| CC | GO:0043204 | Perikaryon | 3/23 | 153/19,594 | 0.0007 | 0.0118 | −0.5773503 |
| MF | GO:0048018 | Receptor ligand activity | 5/20 | 489/18,410 | 0.0001 | 0.0051 | −0.4472136 |
| MF | GO:0030546 | Signaling receptor activator activity | 5/20 | 496/18,410 | 0.0002 | 0.0051 | −0.4472136 |
| MF | GO:0019865 | Immunoglobulin binding | 2/20 | 24/18,410 | 0.0003 | 0.0067 | 1.4142136 |
| KEGG | hsa04640 | Hematopoietic cell lineage | 3/12 | 99/8,164 | 0.0004 | 0.0116 | 1.7320508 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially expressed gene; BP, biological process; CC, cellular component; MF, molecular function.
Link between KLRB1 expression and immune cell infiltration
Additionally, an assessment was conducted to determine if a link existed between KLRB1 expression levels and the infiltration status of immune cells. We used ssGSEA from the R package and R from Spearman to study the potential association between KLRB1 expression levels and 24 immune cell types. The findings indicated a substantial link between KLRB1 expression and all immune cells (Figure 5A). Furthermore, heatmap visualization aided in evaluating and illustrating the varying degrees of correlation among the ratios of 24 distinct tumor-infiltrating immune cell subsets (Figure 5B).
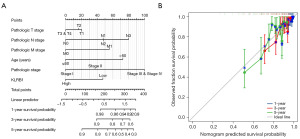
Nomogram development and validation utilizing the independent factors
A nomogram was developed utilizing independent OS-related factors to enable the prognosis prediction of individuals with BRCA. This predictive tool assigns a higher total score to patients with a less favorable prognosis (Figure 6A). To evaluate the prognosis-predictive capacity of the nomogram, calibration curves were employed (Figure 6B), confirming its effectiveness in prognosis prediction.
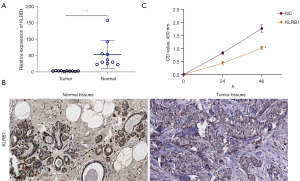
High expression of KLRB1 in BRCA
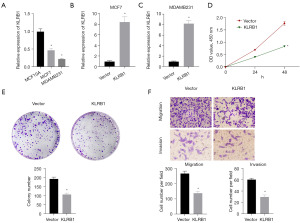
For assessing the potential utility of KLRB1 as a biological marker for BRCA, the expression of KLRB1 in BRCA tissues was further verified using qPCR and IHC. Both qPCR and immunohistochemical outcomes indicated diminished KLRB1 expression in BRCA (Figure 7A,7B). Subsequently, the MCF7 cell line was transfected with a KLRB1-targeted pEZ-M03 vector. CCK8 assay showed that the proliferation of MCF7 cells decreased after transfection with KLRB1 (Figure 7C). Next, we evaluated the expression of KLRB1 in MCF10A cell line, MCF7 cell line and MDAMB231 cell line, and found that the expression of KLRB1 in MCF7 cell line and MDAMB231 cell line was significantly lower than that in MCF10A cell line (Figure 8A). qPCR detection of MCF7 cell lines and MDAMB231 cell lines transfected with KLRB1 targeted pez-m03 vector showed that the expression of KLRB1 in the cell lines transfected with KLRB1 was significantly higher than that in the cell lines transfected with vector (Figure 8B,8C). CCK8 assay and colony formation assay showed that the proliferation of MDAMB231 cell line decreased after transfection of KLRB1 (Figure 8D,8E). Transwell assay showed that the migration and invasion ability of MDAMB231 cell line decreased after transfected with KLRB1 (Figure 8F).
Discussion
BRCA poses a significant threat to the health of women, being the most prevalent cancer and the second major contributor to cancer-related mortality among females (15,16,17). Therefore, there is a pressing need to identify precise biological markers that can facilitate early detection and continuous monitoring of disease progression. As previous research has indicated, the EMC (ER membrane protein complex subunit) is critically involved in the onset and progression of human cancer (18,19). Limited research has explored the link between the expression of KLRB1 and the prognosis of BRCA. This study delved into the potential mechanism governing the role of KLRB1 in promoting BRCA, as well as its feasibility as a potential molecular biological marker.
The comprehensive pan-cancer analysis revealed the consistent downregulation of KLRB1 across various cancer types. Notably, elevated KLRB1 expression correlated with improved OS in individuals with BRCA. Analysis of various clinical stages revealed a substantial correlation between KLRB1 expression and clinical stages. Univariate and multivariate Cox analyses affirmed the independent prognosis-predictive value of KLRB1 in predicting the prognosis of individuals. Collectively, these findings, along with the ROC analysis outcomes, strongly imply that KLRB1 holds promise as a potential prognostic biological marker for individuals with BRCA.
This study revealed a significant inhibitory effect of the KLRB1 gene on cell proliferation in BRCA cells. Simultaneously, this research aims to ascertain the association between the expression of the KLRB1 gene and the clinical prognosis of individuals with BRCA, indicating that individuals with higher KLRB1 expression experience a more favorable prognosis. These findings will serve as a crucial foundation for further investigating the mechanism of the KLRB1 gene in BRCA and identifying potential therapeutic targets.
The results of GSEA suggested that KLRB1-related differential genes were involved in KEGG organic transformation, Reactome organic signaling pathway, NABA restricted factors, KEGG cytokine-cytokine receptor interaction, and KEGG systemic lupus erythematosus pathways. These pathways widely impact cell proliferation, migration, differentiation, and metabolism. In BRCA, olfactory transduction mediated signal transduction ultimately leads to olfactory perception by recognizing odor molecules and activating signal transduction pathways, which also regulates the apoptotic cycle of olfactory sensory neurons in an olfactory receptor-specific manner. A recent study has indicated that certain olfactory receptors exhibit expression in tissues other than the olfactory epithelium, implying their potential for pleiotropic effects (20). In addition, the cytokine-cytokine receptor interaction signaling pathway is also related to BRCA treatment. Various methods can enhance the growth inhibitory and immunostimulatory effects of interferon and interleukin or inhibit the inflammatory and tumor effects of cytokines, thereby treating BRCA (21,22).
Furthermore, the link between KLRB1 expression and the level of immune infiltration in BRCA was investigated utilizing two approaches, ssGSEA and Spearman. KLRB1 exhibited the highest positive correlation with T cells and cytotoxic cells. T cells are a major subclass of lymphocytes, possessing diverse biological functions, including directly targeting and killing specific cells, aiding or inhibiting antibody production by B cells, responding to specific antigens and mitogens, and generating cytokines (23). Research has demonstrated that T cells can directly inhibit BRCA cells and improve the prognosis of BRCA individuals (24,25). Cytotoxic T lymphocyte (CTL) is a specific T cell that secretes various cytokines to participate in immunity and has a strong anti-tumor effect (26). Study has shown that CTL can effectively inhibit BRCA cells and inhibit the onset and angiogenesis of BRCA (27).
Finally, the results were validated by qPCR, IHC, and CCK8 assays. The study demonstrated a significant decrease in KLRB1 expression in corresponding non-tumor tissues. Additionally, the enhanced expression of KLRB1 led to a decrease in the proliferation and invasion capacity of BRCA MCF7 cells. These collective findings highlight that KLRB1 holds promise as a potential predictive tumor marker for individuals with BRCA.
In conclusion, the study aimed to investigate the mechanism and potential therapeutic targets associated with the KLRB1 gene in BRCA. Through the examination of KLRB1 gene expression and function, the aim is to unveil its crucial involvement in the onset and development of BRCA. This endeavor also aspires to offer novel approaches and techniques for addressing the treatment of BRCA. Nonetheless, the precise mechanism via which KLRB1 influences the tumor immune microenvironment and the progression of tumors in BRCA remains to be fully understood. Additional fundamental research and clinical trials are warranted to comprehensively unravel the biological impacts of KLRB1 in BRCA.
Conclusions
In summary, the elevated expression level of KLRB1 is related to prognostic significance and KLRB1 is positively linked to T cells and cytotoxic cells. Consequently, KLRB1, potentially linked to immune infiltration, could serve as a predictive indicator for individuals with BRCA.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1231/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1231/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1231/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-1231/coif). The authors report funding from the Natural Science Foundation of Liaoning province (No. 2019-ZD-1020). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783-91. [Crossref] [PubMed]
- Loibl S, Poortmans P, Morrow M, et al. Breast cancer. Lancet 2021;397:1750-69. [Crossref] [PubMed]
- Coughlin SS. Epidemiology of Breast Cancer in Women. Adv Exp Med Biol 2019;1152:9-29. [Crossref] [PubMed]
- Mathewson ND, Ashenberg O, Tirosh I, et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 2021;184:1281-1298.e26. [Crossref] [PubMed]
- Di W, Fan W, Wu F, et al. Clinical characterization and immunosuppressive regulation of CD161 (KLRB1) in glioma through 916 samples. Cancer Sci 2022;113:756-69. [Crossref] [PubMed]
- Zhou X, Du J, Liu C, et al. A Pan-Cancer Analysis of CD161, a Potential New Immune Checkpoint. Front Immunol 2021;12:688215. [Crossref] [PubMed]
- Zhao H, Yin X, Wang L, et al. Identifying tumour microenvironment-related signature that correlates with prognosis and immunotherapy response in breast cancer. Sci Data 2023;10:119. [Crossref] [PubMed]
- Junjun S, Yangyanqiu W, Jing Z, et al. Prognostic model based on six PD-1 expression and immune infiltration-associated genes predicts survival in breast cancer. Breast Cancer 2022;29:666-76. [Crossref] [PubMed]
- Li W, Guo X, Chen C, et al. The prognostic value of arachidonic acid metabolism in breast cancer by integrated bioinformatics. Lipids Health Dis 2022;21:103. [Crossref] [PubMed]
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938-45. [Crossref] [PubMed]
- Li Y, Gao Y, Niu X, et al. LncRNA BASP1-AS1 interacts with YBX1 to regulate Notch transcription and drives the malignancy of melanoma. Cancer Sci 2021;112:4526-42. [Crossref] [PubMed]
- Li YL, Gao YL, Niu XL, et al. Identification of Subtype-Specific Metastasis-Related Genetic Signatures in Sarcoma. Front Oncol 2020;10:544956. [Crossref] [PubMed]
- Yang Y, Li Y, Qi R, et al. Development and Validation of a Combined Glycolysis and Immune Prognostic Model for Melanoma. Front Immunol 2021;12:711145. [Crossref] [PubMed]
- Krastev T, van Turnhout A, Vriens E, et al. Long-term Follow-up of Autologous Fat Transfer vs Conventional Breast Reconstruction and Association With Cancer Relapse in Patients With Breast Cancer. JAMA Surg 2019;154:56-63. [Crossref] [PubMed]
- Fahad Ullah M. Breast Cancer: Current Perspectives on the Disease Status. Adv Exp Med Biol 2019;1152:51-64. [Crossref] [PubMed]
- Britt KL, Cuzick J, Phillips KA. Key steps for effective breast cancer prevention. Nat Rev Cancer 2020;20:417-36. [Crossref] [PubMed]
- Wang X, Xia Y, Xu C, et al. ER membrane protein complex subunit 6 (EMC6) is a novel tumor suppressor in gastric cancer. BMB Rep 2017;50:411-6. [Crossref] [PubMed]
- Shen MX, Ding JB. Expression levels and roles of EMC-6, Beclin1, and Rab5a in the cervical cancer. Eur Rev Med Pharmacol Sci 2017;21:3038-46. [PubMed]
- Ranzani M, Iyer V, Ibarra-Soria X, et al. Revisiting olfactory receptors as putative drivers of cancer. Wellcome Open Res 2017;2:9. [Crossref] [PubMed]
- Propper DJ, Balkwill FR. Harnessing cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol 2022;19:237-53. [Crossref] [PubMed]
- Conlon KC, Miljkovic MD, Waldmann TA. Cytokines in the Treatment of Cancer. J Interferon Cytokine Res 2019;39:6-21. [Crossref] [PubMed]
- Chapman NM, Boothby MR, Chi H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol 2020;20:55-70. [Crossref] [PubMed]
- Tchou J, Zhao Y, Levine BL, et al. Safety and Efficacy of Intratumoral Injections of Chimeric Antigen Receptor (CAR) T Cells in Metastatic Breast Cancer. Cancer Immunol Res 2017;5:1152-61. [Crossref] [PubMed]
- Sconocchia G, Lanzilli G, Cesarini V, et al. Direct CD32 T-cell cytotoxicity: implications for breast cancer prognosis and treatment. Life Sci Alliance 2022;5:e202201590. [Crossref] [PubMed]
- Reina-Campos M, Scharping NE, Goldrath AW. CD8(+) T cell metabolism in infection and cancer. Nat Rev Immunol 2021;21:718-38. [Crossref] [PubMed]
- Palazon A, Tyrakis PA, Macias D, et al. An HIF-1α/VEGF-A Axis in Cytotoxic T Cells Regulates Tumor Progression. Cancer Cell 2017;32:669-683.e5. [Crossref] [PubMed]


