
RAC1 as a potential pan-cancer diagnostic, prognostic, and immunological biomarker
Highlight box
Key findings
• Ras-related C3 botulinum toxin substrate (RAC1) can serve as a solid tumor biomarker.
What is known and what is new?
• RAC1 is involved in tumor development and immunotherapy in specific tumors.
• Our results provided a new perspective on the role of RAC1 in pan-cancer diagnosis, prognosis, and potential predictive factor regarding immune-check point inhibitors. Moreover, immunohistochemistry (IHC) results showed that liver hepatocellular carcinoma (LIHC) tissues expressed more RAC1-guanosine triphosphate (GTP) than did normal tissues.
What is the implication, and what should change now?
• RAC1 may be both an immunotherapy target and diagnostic and prognostic biomarker.
• Our study also provides a theoretical basis for studying RAC1 in vitro and in vivo, which can enhance our understanding of how RAC1 can be targeted therapeutically, thereby providing a superior immune-based anticancer strategy.
Introduction
Cancer has become a global health crisis, impacting millions of people worldwide, with increasing morbidity and mortality (1). Cancer not only causes suffering to individuals but also imposes a considerable economic burden on society (2). Over the past decade, even with improvements in the diagnosis and treatment of the disease, many cancer patients still have poor prognosis (3). Thus, it is necessary to find new ways to detect and treat cancer early and effectively. The Cancer Genome Atlas (TCGA) and other public databases are continually improving and developing. Whole genome sequencing provides insight into the similarities and differences among different types of cancer and will hopefully lead to the identification of new potential diagnostic, prognostic and therapeutic targets sometimes with agnostic characteristics.
There is evidence suggesting that genetic and epigenetic alterations are involved in the occurrence and development of tumors. Genetic alterations confer selective growth advantages to cancer cells (4). As an epigenetic alteration, DNA methylation can control gene expression without altering the sequence of the DNA (5). Tumor-suppressor genes that are abnormally methylated have been associated with prognoses in esophageal cancer (6). Additionally, immune infiltrating cells in the tumor microenvironment can contribute both to immune escape of tumors and to long-term disease control (7). In recent years, tumor immunotherapy has shown significant efficacy, especially immune checkpoint inhibitors (ICIs) (8-10). However, most patients remain insensitive to immunotherapy. Thus, the identification of new targets that may better stratified cancer about prognosis and or are predictive for immunotherapy/targeted therapies efficacy constitutes an important challenge in this setting.
Ras-related C3 botulinum toxin substrate 1 (RAC1) is a vital member of the Rho GTPase family. Proteins in this group can be transformed between the guanosine diphosphate (GDP)-bound state (inactive) and guanosine triphosphate (GTP)-bound state (active) (11). Many cellular processes are regulated by RAC1, including cell proliferation, apoptosis, invasion and metastasis, and angiogenesis, which promote tumorigenesis and development. RAC1 has also been implicated in cancer therapy resistance (12). Over the past few years, several studies have demonstrated the abnormal expression or activation of RAC1 in some cancers that contributes to poor prognosis (13-15). A relationship between RAC1 and immunity has also been found. RAC1 P29S mutant melanoma cells have a higher level of programmed cell death ligand-1 expression than do RAC1 wild-type cells and can evade immune surveillance (16). In B-cell chronic lymphocytic leukemia, lenalidomide, as an immunomodulatory agent, restores normal levels of RAC1 activity in T cells, thereby maintaining T-cell adhesion and motility (17). Based on these findings, it may be possible to analyse RAC1 expression/alteration for cancer diagnosis, prognosis, and predictive factor for immunotherapy. Thus far, most studies on the RAC1 gene have been limited to specific tumors, and systematic research on its role is lacking. Thus, our study aimed to explore the role of RAC1 in a wider range of solid tumors in order to reveal innovative ideas and methods for clinical studies and treatments in cancer.
In this study, database information was used to analyze RAC1 expression and investigate its prognostic value (Table 1). The level of RAC1 expression was notably associated with pathological stages, RAC1 promoter methylation, immune cell infiltration, immune-related genes, tumor mutational burden, and microsatellite instability. As a use case, we employed gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) to analyze the biological function and pathway of RAC1 in liver hepatocellular carcinoma (LIHC). Based on tumor stage and RAC1 expression, the existing prognostic nomogram was better able to predict patients’ overall survival (OS) rates. Finally, through immunohistochemistry (IHC), we further confirmed the expression of RAC1-GTP in LIHC. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2016/rc).
Table 1
| Tumor | RAC1 Expression | Prognosis (OS) | TAMs |
|---|---|---|---|
| ACC | Higher | Poor | N |
| BLCA | Higher | Poor | N |
| BRCA | Higher | Poor | Positive |
| CESC | Higher | Poor | Positive |
| CHOL | Higher | Better | N |
| COAD | Higher | Poor | N |
| DLBC | Higher | Better | N |
| ESCA | Higher | N | N |
| GBM | Higher | Poor | N |
| HNSC | Higher | Poor | N |
| KICH | N | Poor | N |
| KIRC | Higher | Poor | Positive |
| KIRP | Higher | N | Negative |
| LAML | Lower | N | N |
| LGG | Higher | Poor | Positive |
| LIHC | Higher | Poor | Positive |
| LUAD | Higher | Poor | Positive |
| LUSC | Higher | N | Positive |
| MESO | N | Poor | N |
| OV | Higher | Poor | N |
| PAAD | Higher | Poor | Positive |
| PCPG | N | Better | Positive |
| PRAD | Higher | N | N |
| READ | Higher | N | N |
| SARC | Higher | Poor | Positive |
| STAD | Higher | N | Positive |
| SKCM | Higher | Poor | Positive |
| TGCT | Higher | N | Negative |
| THCA | Higher | N | Positive |
| THYM | Higher | N | Positive |
| UCEC | Higher | N | N |
| UCS | Higher | N | N |
| UVM | N | Poor | N |
RAC1, Ras-related C3 botulinum toxin substrate 1; OS, overall survival; TAM, tumor-associated macrophage; ACC, adrenocortical carcinoma; BLCA, bladder cancer; BRCA, breast cancer; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian cancer; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; STAD, stomach adenocarcinoma; SKCM, skin cutaneous melanoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; N, non-significant.
Methods
RAC1 gene expression pattern
Case information regarding messenger RNA (mRNA) expression and clinical features (18,086 samples from 33 cancers) were obtained from TCGA and the Genotype-Tissue Expression (GTEx) project and downloaded from the website(https://xenabrowser.net/datapages/). The TCGA database contained information on paracancerous tissues, however, due to the absence of corresponding paracancerous tissue data for certain tumors in TCGA, data from both TCGA and GTEx were utilized to analyze and compare the expression levels of RAC1 in cancerous and normal tissues. We transformed the RNA sequencing (RNA-seq) data to transcripts per million (TPM), and a log2 (1 + TPM) transformation was performed. Analysis of tumor and normal tissue expression was performed using t-tests. A clinical dataset from TCGA was analyzed to determine whether the expression of RAC1 affected pathological stages. The “ggplot2” R package (version 4.1.1, The R Foundation of Statistical Computing) was used to analyze and visualize the results.
Gene alteration, copy number alteration (CNA), and RAC1 DNA methylation analysis
cBioPortal database (https://www.cbioportal.org/) is a web platform for analyzing tumor genomic characteristics in the RAC1 gene. We used this database to analyze the types and frequency of RAC1. Pearson rank correlation coefficient was calculated to analyze the relationship between RAC1 expression and CNA and DNA methylation. TCGA samples from the University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN; http://ualcan.path.uab.edu/) were used to compare the promoter methylation level of RAC1 between tumors and normal tissues.
Prognostic value of RAC1 gene
TCGA provided us with survival data of tumor patients (n=9,784). Using routing algorithms, we divided samples into low and high groups based on the optimal cutoff point, OS, disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) were evaluated. The “surv-cutpoint” function within the survminer R package was utilized to identify the optimal split point through a comprehensive evaluation of all potential cut points. Forest plot analysis was conducted using the R package “forestplot”, and R packages “survival” and “survminer” were used to draw survival curves. Log-rank P values, hazard ratios (HRs), and 95% confidence intervals were examined.
Establishment and assessment of the nomogram
To determine the prognostic value of the RAC1 expression level in patients with LIHC, we used an online platform (https://www.home-for-researchers.com/static/index.html#/) to conduct univariate and multivariate Cox regression analysis and develop a nomogram of tumor stage and RAC1 expression. Finally, a correction curve was used to determine whether the nomogram was predictive.
GSEA and GSVA of RAC1 gene in LIHC
Based on GSEA and GSVA, we examined the biological function and significance of RAC1 in LIHC. GSEA was implemented using the R package “clusterProfiler”. A set of gene signatures for the Hallmark pathway was obtained from the Molecular Signatures Database and by using the R package “GSVA”, and we obtained the hallmark pathway scores for LIHC.
Relationship between RAC1 gene expression, immune cell infiltration, and immune-related genes
The Tumor Immune Estimation Resource 2 (TIMER2; http://timer.comp-genomics.org/) offers three distinct modules, namely Immune Association, Cancer Exploration, and Immune Estimation, which facilitate the examination of relationships between immune infiltration and various genetic or clinical attributes.
Analysis of the data revealed there to be a relationship between RAC1 levels and immune cells. Next, our study utilized data on pan-cancer immune cell infiltration obtained from a previous study (18). Furthermore, we examined the relationship between RAC1 and genes associated with immunity. To visualize the data, a heatmap was produced using the “pheatmap” package in R.
Correlational research on RAC1 gene expression and tumor mutational burden, as well as microsatellite instability
Using data derived from TCGA, we examined tumor mutational burden in 33 cancer types. The microsatellite instability data were obtained from a published article (19), and the Pearson rank correlation coefficient was calculated to analyze the relationship between RAC1 expression and tumor mutational burden and microsatellite instability. In order to visualize data, a radar map was generated using the “ggradar” package in R.
Immunohistochemical analysis
The Pathological Center of Ningbo City provided paraffin sections of five pairs of LIHC tissue specimens and adjacent tissue specimens from postoperative patients with LIHC from Department of Radiation Oncology, The Affiliated Lihuili Hospital, Ningbo University between January 2021 to December 2021. RAC1-GTP antibody was purchased from NewEast Biosciences (cat no. 26903; RRID: AB_1961793; NewEast Biosciences, Wuhan, Zhejiang). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of The Affiliated Lihuili Hospital, Ningbo University approved this study (No. KY2022SL322-01). Individual consent for this retrospective analysis was waived. Determine the proportion of positive cells based on sample color and staining intensity, and evaluate the expression intensity of cells. If 0–10% is negative, the score is 0; 11–30% are weakly positive, with a score of 1; 31–60% is positive, with a score of 2; 61–100% are strongly positive, with a score of 3.
Statistical analysis
All statistical analyses were performed in R software (version 4.1.1). Changes in RAC1 expression levels between cancer and normal tissues were assessed through t-tests. Univariate Cox regression analysis was employed to calculate the HR and P value for survival analysis. Kaplan-Meier analysis was utilized to examine the survival duration of patients categorized based on high or low RAC1 expression levels. The Pearson rank correlation coefficient was computed in order to examine the association between the variables. A value of P<0.05 (2-sided) was considered significant for the abovementioned statistical analyses.
Results
Tumor and normal tissue expression of RAC1 gene
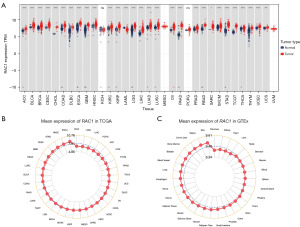
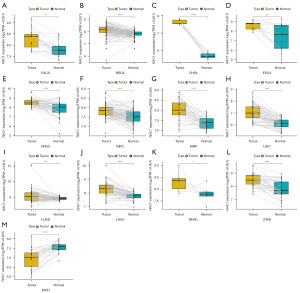
We combined TCGA data with GTEx data to examine RAC1 expression between 33 common cancers and their corresponding normal tissues. A comparison of RAC1 expression between corresponding normal tissues and tumor tissues revealed no significant difference in kidney chromophobe (KICH) or pheochromocytoma and paraganglioma (PCPG), and significant upregulation in other tumor tissues. However, the expression of RAC1 is elevated in normal tissues compared to tumor tissues in acute myeloid leukemia (LAML) (Figure 1A). In 33 tumor tissues from TCGA, esophageal carcinoma (ESCA) exhibited the highest levels of RAC1 and KICH exhibited the lowest levels of RAC1. According to the GTEx database, RAC1 was expressed at the highest level in the skin and the lowest in the pancreas (Figure 1B,1C). TCGA database was used to retrieve paired samples and evaluate RAC1 expression. In tumor tissues from patients with bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), ESCA, head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), LIHC, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), rectum adenocarcinoma (READ), and stomach adenocarcinoma (STAD), RAC1 expression was higher than in adjacent normal tissues. By contrast, its expression was downregulated in KICH (Figure 2).
Relationship between RAC1 gene expression and pathological stages
The expression of RAC1 was analyzed in patients’ various tumors according to the cancer stage to determine if a relationship exists between RAC1 expression and pathological features. The results showed that RAC1 was differentially expressed in different stages of adrenocortical carcinoma (ACC), colon adenocarcinoma (COAD), HNSC, KICH, KIRC, LIHC, LUAD, mesothelioma (MESO), pancreatic adenocarcinoma (PAAD), uterine corpus endometrial carcinoma (UCEC), and ovarian serous cystadenocarcinoma (OV), with the expression level generally being even higher in the more advanced stages (Figure S1).
Gene alteration, CNA, and methylation levels of RAC1 gene promoter
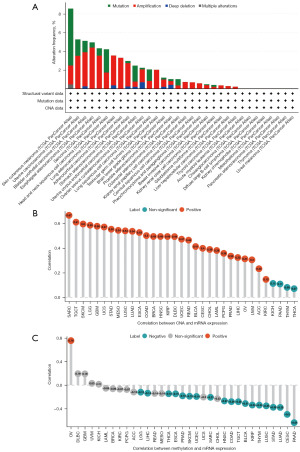
We used the cBioPortal database to study RAC1 gene alteration and CNA. The results showed that RAC1 gene alteration was most common in patients with skin cutaneous melanoma (SKCM), at about an 8.7% incidence. The alteration of the RAC1 gene included mutation, amplification, deep deletion, and multiple alterations. The most common form of gene alteration was amplification. Amplification was also a major alteration in uterine carcinosarcoma (UCS), BLCA, ESCA, sarcoma (SARC), LUAD, ACC, STAD, OV, testicular germ cell tumors (TGCT), brain lower grade glioma (LGG), KIRP, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), PCPG, BRCA, glioblastoma multiforme (GBM), LIHC, and thyroid carcinoma (THCA) (Figure 3A). Additionally, rather than KICH, pancreatic cancer (PANC), thymoma (THYM), and THCA, RAC1 expression correlated positively with CNA (Figure 3B). Furthermore, RAC1 promoter hypermethylation was accompanied by down-regulation of RAC1 expression in LGG, LIHC, THCA, ESCA, prostate adenocarcinoma (PRAD), SKCM, UCEC, SARC, HNSC, COAD, TGCT, BLCA, KIRP, THYM, LUSC, STAD, LUAD, CESC, and PANC tumor tissues; conversely, RAC1 expression increased with hypermethylation of its promoter in OV tumor tissues (Figure 3C).
Next, we investigated the association between RAC1 promoter methylation level and cancers well as its prognostic value. Compared with those of normal tissue, RAC1 promoter methylation levels were significantly decreased in BLCA, BRCA, CESC, CHOL, ESCA, HNSC, UCEC, LIHC, LUAD, LUSC, PRAD, and TGCT tumor tissues. However, the methylation level exhibited a modest increase in KIRP tumor tissues (Figure S2). In order to determine whether RAC1 promoter methylation has prognostic significance, Kaplan-Meier analyses were conducted. The hypermethylation of RAC1 promoter in LIHC, LUAD, PANC, and THYM suggested increased OS but was associated with reduced OS in KIRP. The hypermethylation of RAC1 promoter was a good prognostic factor in patients with LUAD for DSS and in PANC for DFI. Regarding PFI, high RAC1 promoter methylation level in CHOL was significantly associated with better PFI, whereas RAC1 promoter hypermethylation level in KIRP was significantly associated with poor PFI (Figure S3).
Survival analysis of patients with altered RAC1 gene expression
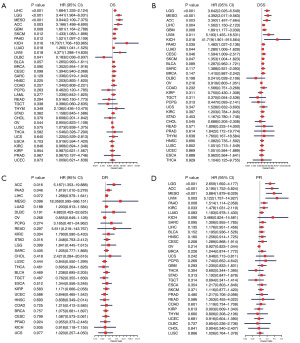
We sought to determine whether RAC1 expression is associated with survival outcomes. Indicators of survival prognosis included OS, DSS, DFI, and PFI. Univariate Cox regression analysis of 33 tumor outcomes indicated that RAC1 expression had a significant correlation with OS in 11 tumors. RAC1 was a risk factor in some tumor types, particularly in KICH. However, it was a protective factor in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) (Figure 4A). The DSS analysis revealed that RAC1 was a risk factor for 11 tumor types, including LGG, MESO, ACC, LIHC, GBM, uveal melanoma (UVM), KICH, PANC, LUAD, CESC, and SKCM (Figure 4B). In the DFI analysis, RAC1 was found to be a risk prognostic factor for ACC and PANC (Figure 4C). In terms of PFI, high RAC1 expression was found to be associated with clinical outcomes of LGG, ACC, MESO, UVM, PANC, and KIRC (Figure 4D).
Kaplan-Meier curves were then generated to determine the prognostic value of RAC1 expression. Results indicated that high RAC1 expression was associated with poor OS in 18 cancer types, including ACC, BLCA, and BRCA, among others. In contrast, the high expression of RAC1 in CHOL, DLBC, and PCPG was associated with better survival (Figure 5). With regards to DSS, the analysis revealed that an increase in RAC1 expression was associated with shorter survival in most cancers, but increased expression of RAC1 in THCA and KIRP was associated with longer DSS (Figure S4).
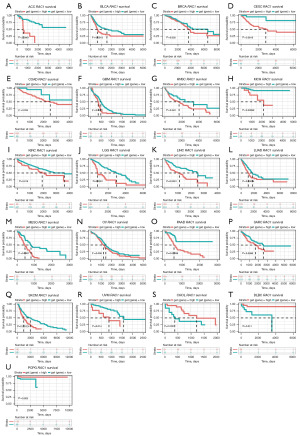
The upregulation of the RAC1 gene was found to be linked to unfavorable DFI in eight cancer types, including ACC, LGG, LIHC, and others, but linked to better DFI in HNSC (Figure S5). Furthermore, RAC1 expression was upregulated in the majority of cancers, which was related to a worse PFI. However, in the case of LUSC, TGCT, THCA, and KIRP, RAC1 upregulated was associated with a better PFI (Figure S6).
Nomogram of LIHC
Due to the former evidences seems to emerge that patients with LIHC who exhibit high expression of RAC1 are at risk of a poor prognosis. Univariate Cox regression analysis was conducted, and tumor size, metastasis sites, and RAC1 were associated with survival and prognosis in LIHC. According to multivariate Cox regression analysis, tumor size and RAC1 were independent risk factors in patients with LIHC (Figure 6A,6B). We then developed a nomogram based on tumor stage and RAC1 expression level to the predict 1-, 3-, and 5-year survival (Figure 6C). In order to determine the accuracy of the prediction model, calibration plots were drawn. The results demonstrated that the nomogram had good predictive ability (Figure 6D).
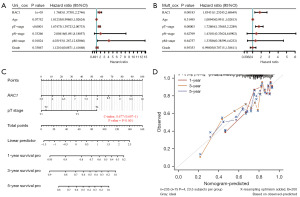
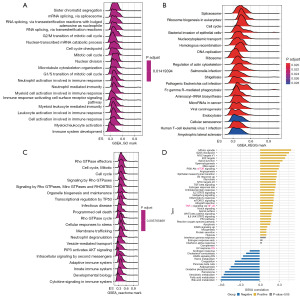
Analysis of GSEA and GSVA in LIHC for RAC1 gene
To investigate the biological function of RAC1, we conducted GSEA on individuals with LIHC using the “clusterProfiler” package in R. The results showed major enrichment in immune regulation and cell cycle pathways, including the neutrophil, myeloid cell, and leukocyte activation involved in the immune response; neutrophil- and myeloid leukocyte-mediated immunity; adaptive and innate immune systems; neutrophil degranulation; cytokine signaling in the immune system, G2/M and G1/S transition of mitotic cell cycle; and cell cycle checkpoint (Figure 7A-7C). Meanwhile, the GSVA results indicated that RAC1 is related to pathways that promote tumor development and immunity, including the mitotic spindle, G2/M checkpoint, DNA repair, PI3K-AKT-mTOR signaling, interleukin 2 (IL-2)-STAT5 signaling, the inflammatory response, and IL-6-JAK-STAT3 signaling (Figure 7D). According to the results of RAC1, this biomarker may serve as an indicator of Immunotherapy sensitivity in LIHC.
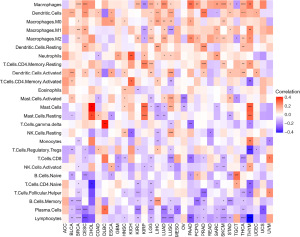
Correlation between RAC1 gene and tumor-associated macrophages (TAMs) infiltration level in tumor microenvironment
Tumor microenvironment immune cells are usually dysfunctional, resulting in immune escape. TAMs are critically involved in tumorigenesis and tumor progression (20). A study has suggested a role for RAC1 in some cancers, but it remains unclear whether abnormal expression of RAC1 has an effect on the infiltration of TAMs (12). To elucidate the role of RAC1 in tumors, we used the TIMER2 database and found a significant positive association between TAM infiltration and RAC1 for most tumors (Figure 8). A published study (18) on immune cell infiltration, also found that the TAM infiltration level was consistent with the RAC1 gene expression (Figure 9).

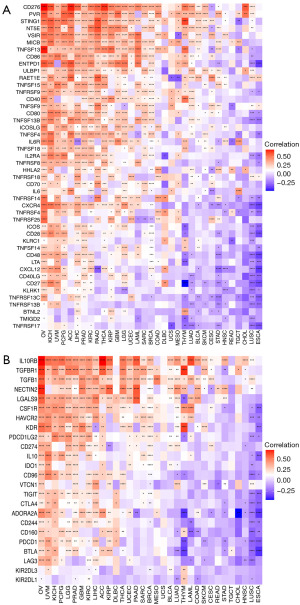
Correlation between RAC1 gene and immune-related genes, tumor mutational burden, and microsatellite instability
We additionally investigated the association of immune-related genes with RAC1. Notably, most genes related to immune stimulation and immunosuppression were positively correlated with RAC1 expression in OV, KICH, UVM, and PCPG. In contrast, in ESCA and LUSC, RAC1 expression showed a negative correlation with many immune stimulation (Figure 10A) and immunosuppression genes (Figure 10B). Among the immune-suppressing genes, IL10RB, TGFBR1, and TGFB1 expression correlated significantly with that of RAC1 in most tumors. Several studies have demonstrated that tumor mutational burden and microsatellite instability are useful biomarkers, which can be helpful in predicting the effectiveness of immunotherapy in cancers (21,22). We examined if RAC1 expression correlated with tumor mutational burden or microsatellite instability. We found that RAC1 expression in MESO, LUAD, and LGG was positively correlated with tumor mutational burden, whereas RAC1 expression in THCA, OV, and COAD was significantly negatively correlated with tumor mutational burden (Figure 11A). Microsatellite instability and RAC1 expression were positively correlated in UVM, MESO, LIHC, and SARC, but negatively correlated in HNSC, LGG, PRAD, and COAD (Figure 11B).
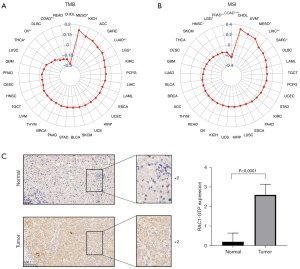
Expression of RAC1-GTP in LIHC and adjacent normal tissues
Further evidence for the role of RAC1 in LIHC was obtained using immunohistochemical analyses performed on postoperative pathological sections from patients with LIHC. The analyses revealed that the expression of RAC1-GTP was higher in LIHC tissues than adjacent normal tissues (Figure 11C).
Discussion
RAC1 participates in a wide range of cellular events, which promote tumorigenesis and development. Abnormal expression or activation of RAC1 is related to inferior prognosis in patients with tumor (13-15), and RAC1 is also involved in tumor microenvironment-mediated immune escape (16,17). The aim of this study was to examine the role of RAC1 in immunotherapy and tumor diagnosis and prognosis. In our literature search, we were unable to find any studies analyzing a full relationship between RAC1 expression in a wide range of cancers. By analyzing whole raw data, similarities and differences between different types of cancer can be identified, thus facilitating the development of personalized cancer prevention and suggest treatment options.
In the study, the expression of RAC1 was examined comprehensively across pan-cancer datasets. In the comparison of tumors with normal tissues, most tumors expressed higher levels of RAC1. Next, we investigated the association between RAC1 expression and pathological stages. For most tumors, RAC1 expression was higher in the advanced/metastatic stages. Furthermore, Cox proportional hazards model and Kaplan-Meier analysis indicated that upregulation of RAC1 expression was correlated with poorer OS, DSS, DFI, and PFI in several cancers, while in other cancers, this was associated with better prognosis. These results suggest that RAC1 could play a role in promoting tumors and could be a biomarker for diagnosis and prognosis.
The chemical modification of DNA, DNA methylation, alters the epigenetic state of a genome without changing its sequence, thus controlling gene expression. Aberrant methylation is a hallmark of cancer progression (5). Currently, promoter methylation is thought to be more easily detectable than that in other locations in genes, which makes it useful for the early detection of cancer (23,24). Our study found that downregulation of RAC1 DNA promoter methylation was accompanied by the upregulation of RAC1 expression in most common cancers. In addition, hypermethylation in some cancers was associated with a good prognosis, but not in KIRP. Therefore, abnormal methylation of RAC1 promoter may affect the development of tumors and the prognosis of patients. These findings suggest that the status of RAC1 DNA promoter methylation could be an early diagnostic marker.
There is evidence supporting the close association of tumor microenvironment with tumorigenesis and the development of tumor. The tumor microenvironment is primarily composed of tumor cells, immune cells, and supporting cells (25). TAMs are immune cells that participate in the immune escape of tumors (7). M2-like TAMs, in particular, as anti-inflammatory cells, play a significant role in promoting various pro-tumorigenic effects in cancer, including regulation of angiogenesis and lymphangiogenesis, immune suppression, induction of hypoxia, promotion of tumor cell proliferation, and facilitation of metastasis (26,27). RAC1 expression levels and prognosis have been found to be closely related in several types of cancer, but the effect of RAC1 on immune cells is unknown. In our study, the association between RAC1 and TAM infiltration was found in a number of tumors. Additionally, the expression of RAC1 was positively correlated with that of immunosuppressive genes including TGFB1 and IL-10. IL-10 promotes the conversion of TAM1 to TAM2, and TAM2 is able to produce TGFB1, which suppresses the antitumor T response (20). The existence of a link between TGFB1, IL-10, and TAM2 further supports the significance of RAC1 in regulating tumor microenvironment. The results suggest a role of RAC1 in the tumor microenvironment, which indicates that it may be an ideal tumor immunotherapy target.
Recently, a breakthrough has been made in immunotherapy in treating some malignant tumors (8-10). However, as immunotherapy has poor efficacy and low response rates for some tumors, only a small portion of patients benefit, limiting its widespread use. Hence, new biomarkers are needed to identify sensitive patients and predict treatment response. Currently, a study has demonstrated that tumor mutational burden and microsatellite instability are useful biomarkers that are helpful in predicting the effectiveness of immunotherapy in cancers (21). High tumor mutational burden and microsatellite instability in patients may indicate a better therapeutic response to ICIs (22,28). In this study, by exploring the relationship between RAC1 expression, tumor mutational burden, and microsatellite instability, we found that RAC1 expression in MESO, LUAD, and LGG was positively correlated with tumor mutational burden. Moreover, an association was observed between microsatellite instability and RAC1 expression in UVM, MESO, LIHC, and SARC. These findings thus point to RAC1 as a potential candidate for predicting the efficacy of immunotherapy for tumors.
Our findings indicate that patients with LIHC who exhibit abnormal expression of RAC1 are at risk of a poor prognosis. To determine the prognostic value of RAC1 among patients with LIHC, a prognostic nomogram was constructed to predict survival. This model had good predictive ability, and will assist clinicians in predicting the prognosis of LIHC so that suitable treatment measures can be administered. The high expression of a protein does not always indicate high activity, as activity level is also related to modification states such as phosphorylation, acetylation, and methylation, as well as the binding of the protein to GTP or GDP. Therefore, studying the activity of a protein by simply detecting the expression level of the total protein provides a limited assessment. At present, no studies exist on RAC1-GTP in LIHC tissues. As shown by IHC, LIHC tissues expressed more RAC1-GTP than did the adjacent normal tissues. The possibility to a relationship between RAC1 expression and immunotherapy sensitivity pave the way to prospective clinical trials in order to find a new predictive factor to atezolizumab activity in this hard-to-treat cancer. But there are also some flaws of this study: (I) in order to understand the true prognostic role of RAC1 we should have had data from an untreated population versus a sample of patients treated with uniform first-line therapy. (II) The final results don’t permit to conclude that RAC1 is an agnostic cancer biomarker due to the controversial results regarding the good or worse prognosis on different tumors. (III) The analysis conducted in this study are derived from raw data and some confounding variables may lead to different biases which may reduce the value of the prognostic and predictive aspect of RAC1. (IV) For these reasons our findings must be confirmed in prospective clinical trials and final conclusion could be considered as a proof of concept for future research.
Conclusions
Our pan-cancer analysis provides insights into the similarities and differences of cancers in relation to RAC1. The findings indicate that RAC1 is a promising biomarker in cancer diagnosis and prognosis and may prove valuable in personalized immunotherapy. However, we used public databases and bioinformatics data, which have limitations. It is necessary to further explore the specific mechanism of RAC1. Future in vivo and in vitro studies of RAC1 may enhance our understanding of how RAC1 can be targeted therapeutically, thereby providing a superior immune-based anticancer strategy.
Acknowledgments
Funding: This work was supported by funding from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2016/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2016/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2016/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2016/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of The Affiliated Lihuili Hospital, Ningbo University (No. KY2022SL322-01). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 2018;103:356-87. [Crossref] [PubMed]
- Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature 2007;446:153-8. [Crossref] [PubMed]
- Mazloumi Z, Farahzadi R, Rafat A, et al. Effect of aberrant DNA methylation on cancer stem cell properties. Exp Mol Pathol 2022;125:104757. [Crossref] [PubMed]
- Liu WJ, Zhao Y, Chen X, et al. Epigenetic modifications in esophageal cancer: An evolving biomarker. Front Genet 2023;13:1087479. [Crossref] [PubMed]
- Pitt JM, Marabelle A, Eggermont A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol 2016;27:1482-92. [Crossref] [PubMed]
- Bagchi S, Yuan R, Engleman EG. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu Rev Pathol 2021;16:223-49. [Crossref] [PubMed]
- Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017;545:495-9. [Crossref] [PubMed]
- Lingel H, Brunner-Weinzierl MC. CTLA-4 (CD152): A versatile receptor for immune-based therapy. Semin Immunol 2019;42:101298. [Crossref] [PubMed]
- Aspenström P. The Role of Fast-Cycling Atypical RHO GTPases in Cancer. Cancers (Basel) 2022;14:1961. [Crossref] [PubMed]
- Liang J, Oyang L, Rao S, et al. Rac1, A Potential Target for Tumor Therapy. Front Oncol 2021;11:674426. [Crossref] [PubMed]
- Li Z, Cao S, Sun Y, et al. TIPE3 is a candidate prognostic biomarker promoting tumor progression via elevating RAC1 in pancreatic cancer. Mol Cancer 2022;21:160. [Crossref] [PubMed]
- Yamaguchi M, Takagi K, Sato A, et al. Rac1 activation in human breast carcinoma as a prognostic factor associated with therapeutic resistance. Breast Cancer 2020;27:919-28. [Crossref] [PubMed]
- Sauzeau V, Beignet J, Bailly C. Rac1 as a Target to Treat Dysfunctions and Cancer of the Bladder. Biomedicines 2022;10:1357. [Crossref] [PubMed]
- Vu HL, Rosenbaum S, Purwin TJ, et al. RAC1 P29S regulates PD-L1 expression in melanoma. Pigment Cell Melanoma Res 2015;28:590-8. [Crossref] [PubMed]
- Ramsay AG, Evans R, Kiaii S, et al. Chronic lymphocytic leukemia cells induce defective LFA-1-directed T-cell motility by altering Rho GTPase signaling that is reversible with lenalidomide. Blood 2013;121:2704-14. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-830.e14. [Crossref] [PubMed]
- Bonneville R, Krook MA, Kautto EA, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol 2017;2017:PO.17.00073.
- Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol 2015;33:445-74. [Crossref] [PubMed]
- Rizzo A, Ricci AD, Brandi G. PD-L1, TMB, MSI, and Other Predictors of Response to Immune Checkpoint Inhibitors in Biliary Tract Cancer. Cancers (Basel) 2021;13:558. [Crossref] [PubMed]
- van Velzen MJM, Derks S, van Grieken NCT, et al. MSI as a predictive factor for treatment outcome of gastroesophageal adenocarcinoma. Cancer Treat Rev 2020;86:102024. [Crossref] [PubMed]
- Linton A, Cheng YY, Griggs K, et al. An RNAi-based screen reveals PLK1, CDK1 and NDC80 as potential therapeutic targets in malignant pleural mesothelioma. Br J Cancer 2018;118:e13. [Crossref] [PubMed]
- Yang X, Dai W, Kwong DL, et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int J Cancer 2015;136:E127-35. [Crossref] [PubMed]
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013;501:346-54. [Crossref] [PubMed]
- Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature 2008;454:436-44. [Crossref] [PubMed]
- Boutilier AJ, Elsawa SF. Macrophage Polarization States in the Tumor Microenvironment. Int J Mol Sci 2021;22:6995. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]


