
A novel PD-1/PD-L1 pathway-related seven-gene signature for the development and validation of the prognosis prediction model for breast cancer
Highlight box
Key findings
• Subtypes of breast cancer (BC) related to the programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) pathway were identified and a gene signature consisting of seven genes related to the PD-1/PD-L1 pathway was determined to predict the prognosis of BC patients.
What is known and what is new?
• The PD-1/PD-L1 pathway is essential for BC.
• We first present PD-1/PD-L1-related subtypes of BC and PD-1/PD-L1-related biomarkers.
What is the implication, and what should change now?
• PD-1/PD-L1 pathway-related biomarkers are closely related to the prognosis of BC patients.
Introduction
Breast cancer (BC/BRCA) is a global health challenge, and is the most common carcinoma in women according to the World Health Organization (WHO) (1). It has been estimated that BC affected one in eight women in 2020 (2). In BC patients with stage IV disease, the average 5-year survival rate is 26% (3). Despite recent advancements in BC treatment and improved patient outcomes, a considerable proportion of patients still do not receive effective therapy (1-3). Therefore, there is an unmet need to identify novel biomarkers for early-stage detection, prognosis, and survival of BC patients.
Recently, the most significant molecular markers associated with BC include estrogen receptor (ER), progesterone receptor (PR), human epidermalgrowth factor receptor-2 (HER2), and the Mib1/Ki-67 proliferation index (4). These markers are firmly established as part of the standard care for all primary, recurrent, and metastatic BC patients (4). Carcinoembryonic antigen (CEA) and carbohydrate antigen (CA) 15-3 antigens are the most valuable serum tumor markers in BC patients (4). The key drawbacks of these markers include their insensitivity to low-volume disease and lack of specific properties (4). Thus, they do not have any utility in the screening or diagnosis of BC (4). Extensive research is currently being conducted on novel biomarkers in BC.
It is commonly known that inhibiting the programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) pathway is essential for enhancing antitumor immune responses in multiple tumor types (5). Indeed, PD-1/PD-L1 immune checkpoint inhibitors (ICIs; PD-1, PD-L1, and CTLA-4 inhibitors) are used for clinical therapy in multiple tumors (6). In BRCA deficiency, type I interferon and proinflammatory cytokines are produced, inducing an innate immune response dependent on STING. The PARP inhibitors are also shown to inhibit GSK3 inactivation and upregulate PD-L1 (7). As a consequence, T-cell activation is suppressed, leading to cancer cell apoptosis (7). Additionally, a number of co-inhibitory immune checkpoint molecules, namely PD-L2, CD47, SIRPα, LAG-3, Tim-3, and TIGIT, have been demonstrated and are universally applicable for assessing immune response (8). Immunotherapy, especially ICIs represented by PD-1 and PD-L1 antibodies, as a new cancer treatment method, has achieved good therapeutic effect in solid tumor patients, including melanoma and lung cancer (9). ICIs may be the preferred choice when aiming for sustained efficacy outcomes, while targeted therapies are primarily considered for patients in need of a relatively rapid objective response. When immunotherapy is administered to melanoma of unknown primary patients, it is likely to result in improved outcomes when contrasted with the melanoma of known primary subset. This may be attributed to their higher immunogenicity, as evidenced by immunologically mediated primary site regression (10). However, in BC, especially triple negative breast cancer (TNBC) with high malignancy, ICI treatment benefits only a small proportion of patients (8,9), indicating that BC limits the efficacy of immunotherapy. In addition, although BC is divided into different types, there are few studies on different types of conservative signaling pathways. The common mechanisms of BC might be the conservative prognostic and predictive factors in BC. As a result, it is of great importance to find out common novel biomarkers and expand the beneficiary population in BC.
Based on the available reports, we acknowledge the significant role of the PD-1/PD-L1 pathway in various tumor types. Nevertheless, research on this topic in relation to BC remains relatively limited. It is crucial to elucidate the association between this pathway and prognosis specifically in BC.
In this study, we performed a systematic investigation to identify prognostic genes in BC patients using The Cancer Genome Atlas (TCGA) sequencing data and focused on assessing its functional roles of PD-1/PD-L1 status in BC patients. We identified two clusters (C1 and C2) of BC patients based on a PD-1/PD-L1 pathway-related signature, and investigated the differences of underlying mechanism, tumor immunity, ferroptosis, and N6-methyladenosine (m6A) expression between different risk groups. Our analysis revealed a notable correlation between the gene signature and prognosis, suggesting its potential as an independent prognostic factor in clinical pathology. We further developed a PD-1/PD-L1 pathway-related gene signature to predict survival of BC patients. Biomarkers related to the PD-1/PD-L1 pathway were associated with a poor prognosis in BC patients. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2270/rc).
Methods
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Datasets of BC patients
We obtained the transcriptome data and the clinical information of 1,101 BRCA patients from TCGA dataset (https://portal.gdc.cancer.gov/projects/TCGA-BRCA). Among the 1,101 BRCA samples, 184 patients were excluded due to a final follow-up of 0, unclear last survival status, or samples with no corresponding expression values.
Identification of prognostic genes in PD-1/PD-L1 pathway
We first analyzed prognostic genes in BC patients using single factor Cox analysis and P values were used to determine the top 20 prognostic genes. Then, these prognostic genes were subjected to functional annotation through Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. The prognostic genes of the PD-1/PD-L1 pathway were identified in BC patients.
Consensus clustering analysis of prognostic genes of the PD-1/PD-L1 pathway
We used the R package ConsensusClusterPlus (version 1.54.0) to cluster consensus data by size, and a maximum of six clusters were created. In order to plot the heatmap, the pheatmap R package (version 1.0.12) was used. The R package survival was used to draw and analyze Kaplan-Meier (KM) curves.
Identification of differentially expressed genes (DEGs) and functional enrichment analysis
DEGs between C1 and C2 in the TCGA-BC patients were screened by “DESeq2” R package based on the criteria [|log fold change (FC)| >1 and P<0.05]. Displaying DEGs was done using volcano plots. An R package called pheatmap (version 1.0.12) was used to create heatmaps for the 50 most significant DEGs. We used the R package ClusterProfiler to perform the Gene Ontology (GO) and KEGG analysis.
Comparative analysis of ferroptosis and m6A between BC subtypes
We obtained ferroptosis-related genes (11) and m6A-related genes (12) from previous research. We analyzed the expression profiles of ferroptosis-related genes and m6A-related genes between C1 and C2 in BC patients. A heatmap of ferroptosis-related genes and m6A-related genes was plotted using pheatmap R package (version 1.0.12). Statistical analyses were performed using R software (version 4.0.3, 2020; the R Foundation for Statistical Computing, Vienna, Austria).
Development of a prognostic gene model in the TCGA-BC cohort
To construct a prognostic model, we used the least absolute shrinkage and selection operator (LASSO) Cox regression method implemented in the “glmnet” R package. The variables with non-zero coefficients were selected to build the prognostic model based on the minimum lambda condition. Each gene’s expression levels were combined by multiplying them by their respective coefficients to calculate a risk score. The median risk score was used to classify TCGA-BC patients into high- and low-risk groups. The R package “survival” was used to draw and analyze KM curves. According to Cox proportional hazards analysis, hazard ratios (HRs) and 95% confidence intervals (CIs) are calculated. An area under the curve (AUC) greater than 0.70 indicates good predictive ability; between 0.70 and 0.50 indicates lower accuracy; and less than 0.50 indicates no predictive ability.
Immunohistochemical analysis
Based on the transcript levels of genes of the PD-1/PD-L1 pathway, we further investigated the protein expression of these genes between BC tissues and normal tissues using the Human Protein Atlas (HPA) database (https://www.proteinatlas.org).
Statistical analysis
We used R (version 3.6.3) for our statistical analysis. Prognostic factors for BC were found using survival analysis at a significance level of P<0.05. Log-rank tests were used to evaluate the comparison between high-risk and low-risk groups based on KM survival analysis. The statistical significance level was set at a two-sided P value of 0.05.
Results
KEGG pathways of prognostic genes in BC
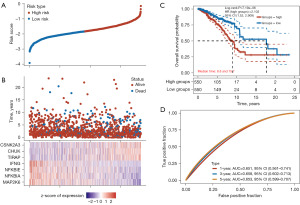
To investigate the mechanism of prognostic genes in BC patients, 866 prognostic genes were found using the single factor Cox analysis and the top 20 genes are shown in Figure 1A based on P value (table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-1.xlsx).
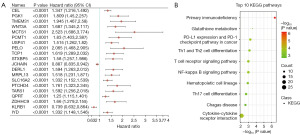
KEGG enrichment analyses were carried out based on these genes. Interestingly, the pathways of prognostic genes were mainly associated with primary immunodeficiency, cytokine-cytokine receptor interaction, and PD-L1 expression and PD-1 checkpoint pathway in cancer (Figure 1B and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-2.xlsx), which suggested that immune pathway might play an important role in BC. As the PD-1/PD-L1 pathway is closely associated with cancer, we next focused on assessing its functional roles of PD-1/PD-L1 status in BC patients.
Tumor classification based on the PD-1/PD-L1 pathway in BC
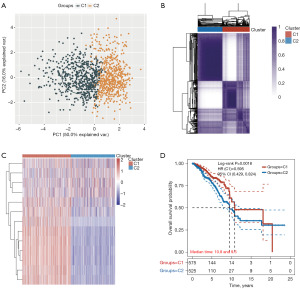
Next, we investigated whether the expression of 13 genes of the PD-1/PD-L1 pathway [including mitogen-activated protein kinase kinase 6 (MAP2K6), NF-kappa-B inhibitor alpha (NFKBIA), CD3D, CD3E, NFKB Inhibitor Epsilon (NFKBIE), Interferon gamma (IFNG), CD247, ZAP70, Toll/interleukin-1 receptor domain-containing adapter protein (TIRAP), PDCD1, CD3G, IkappaB kinase (CHUK), and Casein kinase 2 alpha 3 gene (CSNK2A3)] was related to BC subtypes. TCGA-BC patients were divided into different subtypes using these genes through consistency cluster analysis. When the clustering variable (kappa) was 2, two clusters of TCGA-BC patients were well constructed based on the results of principal component analysis (PCA) and consistency clustering matrix analysis (Figure 2A,2B and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-3.xlsx). In addition, two clusters of TCGA-BC patients were well separated, as displayed in Figure 2C. We further compared overall survival (OS) times between two groups. KM curves of OS showed that patients in C1 had better survival than those in C2 (HR =0.595; 95% CI: 0.429–0.824; P=0.0018, Figure 2D and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-3.xlsx).
Functional annotation between two clusters
Further exploration of that underlying mechanism was required between these two clusters in BC patients. According to the volcano plot, we identified 372 DEGs (367 upregulated; 5 downregulated) between C1 and C2 using the cut-off criteria (FC ≥2 or FC ≤0.5, and P value <0.05) (Figure 3A and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-3.xlsx). Several upregulated genes (CD3E, CD3D, LCK, CD2, etc.) were displayed by volcano plot (Figure 3A). The top 50 significantly DEGs were shown in the heatmap, indicating different characteristics between the two clusters (Figure 3B).
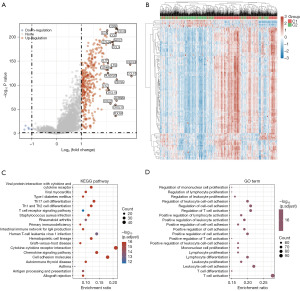
Functional annotation between the two clusters was analyzed based on the information of these 372 DEGs. KEGG enrichment analyses revealed that the pathways were primarily related to viral protein interaction with cytokine and cytokine receptor, Th1 and Th2 cell differentiation, and primary immunodeficiency (Figure 3C and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-4.xlsx). Moreover, GO enrichment analyses showed a similar result, with DEGs being mainly associated with regulation of immune cell proliferation, regulation of T cell activation, and positive regulation of cell-cell adhesion (Figure 3D and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-4.xlsx). Thus, these results further suggested that tumor classification based on the PD-1/PD-L1 pathway in BC was mainly related to immune-related biological processes and signaling pathways. This result further indicated that C1 patients might exhibit increased immune cell proliferation and positive regulation of T cell activation.
Comparative analysis of ferroptosis and m6A between BC subtypes
It has been reported that anti-PD-1/PD-L1 resistance induces tumor cell ferroptosis (5,13), indicating that the PD-1/PD-L1 pathway has a crucial role in this process. Thus, we analyzed the expression profiles of ferroptosis-related genes between C1 and C2 in BC patients. Heatmaps and boxplots depicted that 8 of the 25 ferroptosis-related genes were significantly different between the two clusters, including EMC2, NFE2L2, HSPB1, SLC1A5, SAT1, GLS2, DPP4, and ACSL4 (Figure 4A,4B and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-5.xlsx). These results may suggest that the PD-1/PD-L1 pathway and ferroptosis are correlated with BC prognosis.
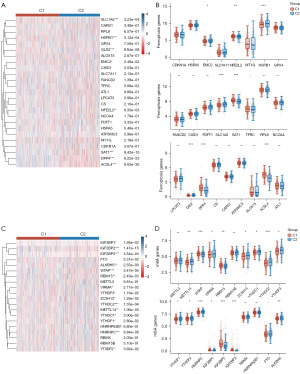
Recently, it has been shown that m6A demethylases fat mass and obesity-associated (FTO) protein promotes PD-L1 expression through inducing m6A demethylation in cancers (14). We further analyzed the expression profiles of m6A-related genes between C1 and C2 in BC patients (Figure 4C,4D and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-4.xlsx). Interestingly, heatmaps and boxplots depicted that 15 of the 20 m6A-related genes were significantly different between the two clusters, including METTL14, WTAP, VIRMA, RBM15, ZC3H13, YTHDC1, YTHDC2, YTHDF3, YTHDF1, YTHDF2, HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, and ALKBH5 (Figure 4A,4B and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-6.xlsx). In summary, we found a correlation between the PD-1/PD-L1 pathway and m6A-related gene expression in BC.
Development of a prognostic gene model in the TCGA-BC cohort
To explore the prognostic value of 13 genes of the PD-1/PD-L1 pathway, we developed a PD-1/PD-L1 pathway-related prognostic model to predict the clinical prognosis. The LASSO regression algorithm was used to further optimize these genes, yielding seven genes of the PD-1/PD-L1 pathway associated with TCGA-BC prognosis, including MAP2K6, NFKBIA, NFKBIE, IFNG, TIRAP, CHUK, and CSNK2A3 (Figure 5A and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-5.xlsx). We calculated risk score as follows: risk score = (−0.3866 × MAP2K6 expression) + (−0.3106 × NFKBIA expression) + (−0.0521 × NFKBIE expression) + (−0.1587 × IFNG expression) + (0.207 × TIRAP expression) + (0.2416 × CHUK expression) + (0.2465 × CSNK2A3 expression). The cut-off points for the median risk score categorized the BC patients into two groups (high-risk vs. low-risk; Figure 5B). Based on KM curves, the low-risk group was significantly more likely to survive than the high-risk group (P=7.19e−06, Figure 5C). The receiver operating characteristic (ROC) curve analyses were used to assess the prognostic value of these seven genes. AUCs for OS at 1, 3, and 5 years were 0.651, 0.658, and 0.653, respectively (Figure 5D and table available at https://cdn.amegroups.cn/static/public/tcr-23-2270-7.xlsx).
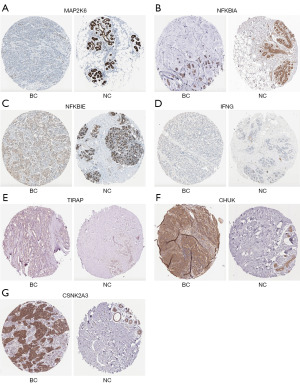
Immunohistochemical analysis of seven genes between BC tissues and normal tissues
Low-risk individuals had significantly higher levels of transcript levels for MAP2K6, NFKBIA, NFKBIE, and IFNG than high-risk individuals. As compared to the high-risk group, the low-risk group showed significantly lower levels of TIRAP, CHUK, and CSNK2A3. We used the HPA database to analyze the protein expression of these genes. The immunohistochemical expression of these genes showed that the expression of MAP2K6, NFKBIA, and NFKBIE was low in BC tissues, with the exception of IFNG (Figure 6A-6D), whereas TIRAP, CHUK, and CSNK2A3 were overexpressed in BC tissues (Figure 6E-6G). These results suggested that the protein levels of six genes were consistent with their transcript levels.
Discussion
In this study, we first studied prognostic genes in BC patients using TCGA data and found that the PD-1/PD-L1 pathway mainly associated with the BC patients. A low-risk C1 group and a high-risk C2 group of BC patients were constructed based on a PD-1/PD-L1 pathway-related signature. The functional analyses suggested that the DEGs between these groups were mainly associated with immune-related pathways. We also found that ferroptosis and m6A between low- and high-risk groups were significantly different. We further developed a PD-1/PD-L1 pathway-related gene signature to predict survival of BC patients and identified seven genes (MAP2K6, NFKBIA, NFKBIE, IFNG, TIRAP, CHUK, and CSNK2A3). Taken together, PD-1/PD-L1 pathway-related biomarkers strongly correlated with the prognosis of BC patients.
According to KEGG enrichment analyses of prognostic genes, primary immunodeficiency and the PD-1/PD-L1 pathway were closely related to BC, indicating that the tumor immune pathway contributes to occurrence of BC. Recently, ICIs (PD-1, PD-L1, and CTLA-4 inhibitors) have been used for clinical therapy in multiple tumors (6). In addition, various co-inhibitory immune checkpoint molecules, such as PD-L2, CD47, SIRPα, LAG-3, Tim-3, and TIGIT, have been validated as universally applicable for assessing immune response (8,15). However, in BC, ICI treatment benefits a small proportion of patients (15-17), indicating that BC limits the efficacy of immunotherapy and requires new novel targets.
Recently, some important molecular markers associated with BC include ER, PR, HER2, and the Mib1/Ki-67 proliferation index (4). These biomarkers are firmly established as part of the standard care for all primary, recurrent, and metastatic in BC (4). In addition, the prognostic biomarkers of some factors, including tumor microenvironment, tumor-specific ceRNA, and lncRNA, were identified based on the prognosis prediction models (18-21).
Functional annotation between the low-risk C1 group and high-risk C2 group of BC patients further confirmed that immune-related pathways (cytokine and cytokine receptor and Th1 and Th2 cell differentiation) were associated with BC. We further found a correlation of the PD-1/PD-L1 pathway with ferroptosis- and m6A-related gene expression for different risk group in BC. Therefore, these results further suggested that tumor classification based on the PD-1/PD-L1 pathway could differentiate BC.
Due to the strong correlation between status of the PD-1/PD-L1 pathway and clinical prognosis in BC, high-risk and low-risk patients were divided using a gene signature. A seven gene signature was identified using LASSO regression analysis, comprising MAP2K6, NFKBIA, NFKBIE, IFNG, TIRAP, CHUK, and CSNK2A3, displayed a substantial effect on survival prediction. In these genes, TIRAP, CHUK, and CSNK2A3 were overexpressed in BC tissues compared with normal tissues, whereas the expression of MAP2K6, NFKBIA, and NFKBIE was low in BC tissues, with the exception of IFNG, at the protein level.
TIRAP has been shown to regulate cytokine secretion and the inflammatory response through TLR2/TLR4 signaling in BC (22). CHUK, encoding inhibitor of nuclear factor kappa-B kinase subunit, plays an essential role in the NF-kappa-B signaling pathway in BC (23). CSNK2A3 plays a role in the pathogenesis of the lung cancer development and progression (24). Dual specificity MAP2K6 is an essential component of the Mitogen-activated protein kinase (MAPK) pathway in many cancers (25,26). NFKBIA and NFKBIE is involved in sporadic BC (27).
The study is subject to several limitations. Despite conducting a comprehensive evaluation and analysis using multiple platforms and databases, the conclusions are solely based on in vitro analysis. Due to the inherent limitations of computational analysis, additional network experiments are necessary to elucidate the underlying mechanisms. Therefore, further experimental exploration is necessary to uncover the potential mechanisms of these crucial genes.
Taken together, the expression of gene signatures related to these pathways predicted BC patients’ prognoses. A certain degree of clinical satisfaction can be achieved with this feature in terms of BC management.
Conclusions
PD-1/PD-L1 pathway-related subtypes of BC were identified, which were closely associated with the immune microenvironment, the ferroptosis status, and m6A in BC patients. The gene signature involved in the PD-1/PD-L1 pathway might help to make a distinction and predict prognosis in BC patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2270/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2270/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2270/coif). S.B. is from AELIA Organization. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gallardo M, Kemmerling U, Aguayo F, et al. Curcumin rescues breast cells from epithelial mesenchymal transition and invasion induced by anti miR 34a. Int J Oncol 2020;56:480-93. [PubMed]
- Li H, Li L, Xue C, et al. A Novel Ferroptosis-Related Gene Signature Predicts Overall Survival of Breast Cancer Patients. Biology (Basel) 2021;10:151. [Crossref] [PubMed]
- Peric L, Vukadin S, Petrovic A, et al. Glycosylation Alterations in Cancer Cells, Prognostic Value of Glycan Biomarkers and Their Potential as Novel Therapeutic Targets in Breast Cancer. Biomedicines 2022;10:3265. [Crossref] [PubMed]
- Tarighati E, Keivan H, Mahani H. A review of prognostic and predictive biomarkers in breast cancer. Clin Exp Med 2023;23:1-16. [PubMed]
- Jiang Z, Lim SO, Yan M, et al. TYRO3 induces anti-PD-1/PD-L1 therapy resistance by limiting innate immunity and tumoral ferroptosis. J Clin Invest 2021;131:e139434. [Crossref] [PubMed]
- Yi M, Zheng X, Niu M, et al. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer 2022;21:28. [Crossref] [PubMed]
- Revythis A, Limbu A, Mikropoulos C, et al. Recent Insights into PARP and Immuno-Checkpoint Inhibitors in Epithelial Ovarian Cancer. Int J Environ Res Public Health 2022;19:8577. [Crossref] [PubMed]
- Greisen SR, Aspari M, Deleuran B. Co-Inhibitory Molecules - Their Role in Health and Autoimmunity; Highlighted by Immune Related Adverse Events. Front Immunol 2022;13:883733. [Crossref] [PubMed]
- Wojtukiewicz MZ, Rek MM, Karpowicz K, et al. Inhibitors of immune checkpoints-PD-1, PD-L1, CTLA-4-new opportunities for cancer patients and a new challenge for internists and general practitioners. Cancer Metastasis Rev 2021;40:949-82. [Crossref] [PubMed]
- Boussios S, Rassy E, Samartzis E, et al. Melanoma of unknown primary: New perspectives for an old story. Crit Rev Oncol Hematol 2021;158:103208. [Crossref] [PubMed]
- Liu Z, Zhao Q, Zuo ZX, et al. Systematic Analysis of the Aberrances and Functional Implications of Ferroptosis in Cancer. iScience 2020;23:101302. [Crossref] [PubMed]
- Li Y, Xiao J, Bai J, et al. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer 2019;18:137. [Crossref] [PubMed]
- Wang W, Green M, Choi JE, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270-4. [Crossref] [PubMed]
- Liu Z, Yu X, Xu L, et al. Current insight into the regulation of PD-L1 in cancer. Exp Hematol Oncol 2022;11:44. [Crossref] [PubMed]
- Kunkler IH, Williams LJ, Jack WJL, et al. Breast-Conserving Surgery with or without Irradiation in Early Breast Cancer. N Engl J Med 2023;388:585-94. [Crossref] [PubMed]
- Sarhangi N, Hajjari S, Heydari SF, et al. Breast cancer in the era of precision medicine. Mol Biol Rep 2022;49:10023-37. [Crossref] [PubMed]
- Bodewes FTH, van Asselt AA, Dorrius MD, et al. Mammographic breast density and the risk of breast cancer: A systematic review and meta-analysis. Breast 2022;66:62-8. [Crossref] [PubMed]
- Zhong S, Jia Z, Zhang H, et al. Identification and validation of tumor microenvironment-related prognostic biomarkers in breast cancer. Transl Cancer Res 2021;10:4355-64. [Crossref] [PubMed]
- Zheng M, Wu L, Xiao R, et al. Integrated analysis of coexpression and a tumor-specific ceRNA network revealed a potential prognostic biomarker in breast cancer. Transl Cancer Res 2023;12:949-64. [Crossref] [PubMed]
- Wang X, Li X, Jiang W. High expression of RTN4IP1 predicts adverse prognosis for patients with breast cancer. Transl Cancer Res 2023;12:859-72. [Crossref] [PubMed]
- Xu M, Chen Z, Lin B, et al. A seven-lncRNA signature for predicting prognosis in breast carcinoma. Transl Cancer Res 2021;10:4033-46. [Crossref] [PubMed]
- Wang J, Dong B, Tan Y, et al. A study on the immunomodulation of polysaccharopeptide through the TLR4-TIRAP/MAL-MyD88 signaling pathway in PBMCs from breast cancer patients. Immunopharmacol Immunotoxicol 2013;35:497-504. [Crossref] [PubMed]
- Espinoza-Sánchez NA, Győrffy B, Fuentes-Pananá EM, et al. Differential impact of classical and non-canonical NF-κB pathway-related gene expression on the survival of breast cancer patients. J Cancer 2019;10:5191-211. [Crossref] [PubMed]
- Hung MS, Lin YC, Mao JH, et al. Functional polymorphism of the CK2alpha intronless gene plays oncogenic roles in lung cancer. PLoS One 2010;5:e11418. [Crossref] [PubMed]
- Lin S, Liu K, Zhang Y, et al. Pharmacological targeting of p38 MAP-Kinase 6 (MAP2K6) inhibits the growth of esophageal adenocarcinoma. Cell Signal 2018;51:222-32. [Crossref] [PubMed]
- Li Z, Fu J, Li N, et al. Quantitative proteome analysis identifies MAP2K6 as potential regulator of LIFR-induced radioresistance in nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 2018;505:274-81. [Crossref] [PubMed]
- Curran JE, Weinstein SR, Griffiths LR. Polymorphic variants of NFKB1 and its inhibitory protein NFKBIA, and their involvement in sporadic breast cancer. Cancer Lett 2002;188:103-7. [Crossref] [PubMed]


